Hospitalsfaceastarkrealityinmedicalimagingwherelabeleddataarescarceanddomainsdivergewildlyacrosscenters. Across scanners, protocols, and patient cohorts, the visual look of the same anatomy can shift just enough to trip up segmentation systems trained under tidy lab assumptions. A new training strategy—Dual-supervised Asymmetric Co-training (DAC), developed by Assistant Professor Zhao Na and collaborators and published in IEEE Transactions on Multimedia—tackles this friction head-on in cross-domain semi-supervised domain generalization (CD-SSDG). The premise is pragmatic: start with a small labeled set from one source and a large, diverse pool of unlabeled images from elsewhere, then teach a model to generalize without incurring any extra cost at inference. Reported gains concentrate where clinical systems tend to struggle most: small, faint, or low-contrast structures that often dictate outcomes yet evade consistent delineation.
clinical data reality and the cd-ssdg setting
Clinical archives resemble patchwork more than curated corpora modest batch of expert-annotated images from a primary scanner sits alongside large stores of unlabeled images collected over time, frequently under shifting protocols, resolutions, and noise regimes. This asymmetry defines CD-SSDG, a setting that combines two stubborn hurdles—domain mismatch and label scarcity—and exposes fragilities in conventional training recipes. Standard semi-supervised approaches lean on pseudo-labels generated by a model trained on the labeled subset, which works when unlabeled images “look” similar. Under shift, those pseudo-labels wobble, and the training loop can amplify early mistakes, entrenching bias rather than dissolving it.
In parallel, domain generalization (DG) methods promise robustness across centers but often presume multiple labeled source domains or sidestep semi-supervised realities. CD-SSDG is a stricter test: it pairs a single labeled source with many unlabeled, mismatched targets. The result is a realistic yet unforgiving training regime that mirrors hospital constraints and exposes brittle assumptions about i.i.d. data. Segmentation suffers most on anatomies that are small or weakly contrasted, where a slight change in intensity or texture may swamp shape cues. This is precisely where a method must prioritize domain-invariant structure—boundaries, topology, geometry—over appearance quirks to avoid cascading errors across the unlabeled pool.
how dac rethinks co-training to survive domain shift
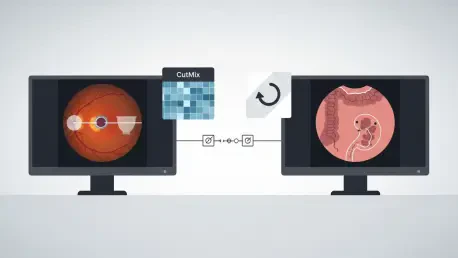
DAC embraces co-training’s intuition—use two learners to cross-check each other—but revises the mechanics to resist domain noise and brittle pseudo-labels. Its first change is dual supervision: beyond exchanging mask-level pseudo-labels on unlabeled images, the two sub-models align in feature space, nudging representations toward domain-invariant cues such as shape, relative layout, and topology. This feature-level agreement acts like a stabilizer, helping the pair keep focus on structure even when pixel intensities drift across scanners. The second change is deliberate asymmetry via auxiliary self-supervised tasks: one learner localizes a mixed patch in CutMix images, the other predicts patch rotation, ensuring they develop complementary perspectives rather than collapsing onto the same bias.
Crucially, these additions live solely in training. The auxiliary heads are simple—one linear layer each—and are discarded at deployment, and the feature-consistency constraint vanishes as well. Inference cost therefore matches a standard single-model backbone, a significant advantage in clinical workflows bound by latency and memory ceilings. The framework drops into common backbones like ResNet-DeepLabv3+ and works with standard optimizers, so it functions as a training recipe rather than a bespoke architecture. By maintaining diversity between sub-models while anchoring them with feature-level guidance, DAC steadily upgrades pseudo-label quality across the unlabeled pool, decoupling core anatomy from domain-specific appearance that otherwise derails learning under shift.
evidence across benchmarks and what it means for deployment
Empirical tests spanned three representative challenges: retinal fundus images for optic disc and optic cup segmentation, colorectal polyp datasets with varied scopes and lighting, and SCGM MRI targeting fine gray matter delineation. Across these benchmarks, DAC outperformed strong semi-supervised and DG baselines, with the most pronounced gains on small, low-contrast targets such as the optic cup. The method remained steady as the labeled fraction shrank—down to a tenth in some settings—suggesting its training signals resist collapse when annotations are scarce. Reported training speed also compared favorably to a leading co-training baseline, likely due to the stabilizing effect of feature alignment and the efficiency of the auxiliary heads.
For deployment, the absence of inference overhead matters as much as raw accuracy. Hospitals commonly face throughput constraints, strict validation protocols, and reproducibility requirements tied to inference behavior. Because DAC’s extra supervision appears only during training, the deployed model behaves like a standard backbone: no additional memory spikes, no ensemble calls, and no latency penalties. This streamlines integration with existing toolchains and reduces friction in audits that scrutinize runtime characteristics. The plug-in nature also shortens the path from lab to clinic, enabling teams to trial DAC with familiar architectures while leveraging the large unlabeled pools that accumulate in routine practice.
limits, next steps, and broader relevance
No method closes every gap. DAC struggled in fundus scenes with multiple vessels crossing the optic disc, where tangled boundaries and local ambiguities undermined both pixel- and feature-level cues. Extremely low-contrast cases posed a similar challenge, as targets nearly melted into background, thinning the structural signals that DAC relies on to stabilize learning. The authors proposed concrete extensions: vessel-aware augmentation to better model intersecting vasculature, and adaptive, multi-view representations that blend multi-scale and frequency-domain cues to sharpen boundaries and capture complementary evidence beyond spatial features alone.
The broader playbook extends beyond the reported datasets. Feature-level supervision, asymmetric co-training, and training-only auxiliaries map naturally to tumor imaging and other high-stakes targets where annotations are costly and boundary precision drives decisions. The field appears to be drifting from brittle pixel-wise pseudo-label dependence toward richer agreement in feature space, and from heavier inference-time ensembles toward lightweight training-time strategies that leave deployment unchanged. In practical terms, the findings encouraged teams to treat DAC as a drop-in training upgrade: exploit unlabeled pools despite domain mismatch, prioritize domain-invariant structure, and reserve complexity for where it pays off—during training rather than at the bedside.